Modelling Ocean Microbes using Satellites
Posted on July 18, 2025 • 3 minutes • 435 words
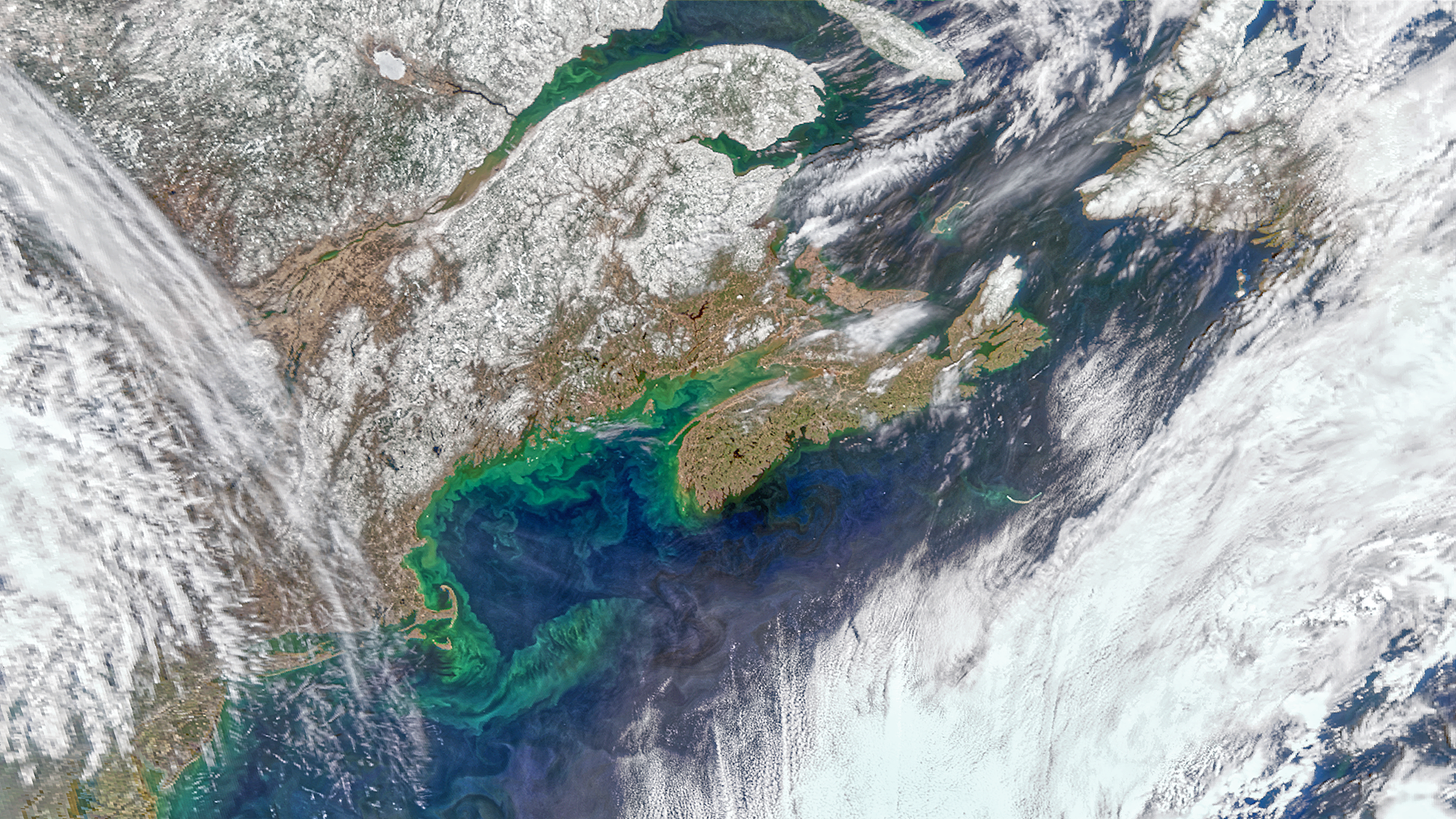
Satellite oceanography has revolutionized our ability to study phytoplankton communities and organic matter in the oceans across large spatial scales and time, yet there remains a gap in the ability to monitor and predict the diversity of the microbial communities that underpin ocean food webs using satellite observational tools. This matters because experimental and observational studies indicate that the source, quality, and quantity of organic matter influences taxonomic and functional diversity changes of microbial communities, which in turn affects biogeochemical cycling, so the strength of this correlation needs to be better understood for modeling responses to climate change.
 We have teamed up with the Marine Optics Lab at Bigelow, lead by Dr. Catherine Mitchell, to augment a long-running NASA satellite ocean color validation time series in a rapidly changing ecosystem – the Gulf of Maine North Atlantic Time Series (GNATS) – with microbial diversity assays to explore the relationship between microbial community structure with phytoplankton and carbon observations. We aim to predict microbial community structure from satellite observational data, and to determine the key variables driving community variation. If successful, this will revolutionize the predictive power of satellite oceanography.
We have teamed up with the Marine Optics Lab at Bigelow, lead by Dr. Catherine Mitchell, to augment a long-running NASA satellite ocean color validation time series in a rapidly changing ecosystem – the Gulf of Maine North Atlantic Time Series (GNATS) – with microbial diversity assays to explore the relationship between microbial community structure with phytoplankton and carbon observations. We aim to predict microbial community structure from satellite observational data, and to determine the key variables driving community variation. If successful, this will revolutionize the predictive power of satellite oceanography.
The question driving our research is: How is microbial community activity and diversity influenced by colored dissolved organic matter (CDOM) and phytoplankton community composition across spatial and temporal scales in coastal shelf ecosystems?
 We hypothesize that: (1) Dominant microbial taxa, functional genes and actively transcribed genes will differ based on the organic matter content of dynamic biogeochemical environments of the Gulf of Maine coastal shelf ecosystem. And (2) Changes in the Gulf of Maine have and will continue to cause temporal and spatial shifts in the taxonomic diversity and activity of microbial communities. Economically significant coastal shelf ecosystems are heterogeneous and sensitive to change, requiring information across both time and space to understand microbial community diversity, dynamics and activity. Yet, traditional nucleic sequencing techniques used to characterize microbial communities are limited in the extent of spatial and temporal observations that can be made. To expand such observations across the entire Gulf of Maine region, we will employ in situ sampling over the course of a year across the GNATS transect that samples different water masses within the Gulf of Maine. We will then integrate observations of microbial community diversity and activity with satellite-derived information on phytoplankton and CDOM to understand the relationship between microbial community diversity and satellite-derived measurements. With this information in hand, we will construct models that predict aspects of microbial community diversity and function using satellite-derived data across the Gulf of Maine.
We hypothesize that: (1) Dominant microbial taxa, functional genes and actively transcribed genes will differ based on the organic matter content of dynamic biogeochemical environments of the Gulf of Maine coastal shelf ecosystem. And (2) Changes in the Gulf of Maine have and will continue to cause temporal and spatial shifts in the taxonomic diversity and activity of microbial communities. Economically significant coastal shelf ecosystems are heterogeneous and sensitive to change, requiring information across both time and space to understand microbial community diversity, dynamics and activity. Yet, traditional nucleic sequencing techniques used to characterize microbial communities are limited in the extent of spatial and temporal observations that can be made. To expand such observations across the entire Gulf of Maine region, we will employ in situ sampling over the course of a year across the GNATS transect that samples different water masses within the Gulf of Maine. We will then integrate observations of microbial community diversity and activity with satellite-derived information on phytoplankton and CDOM to understand the relationship between microbial community diversity and satellite-derived measurements. With this information in hand, we will construct models that predict aspects of microbial community diversity and function using satellite-derived data across the Gulf of Maine.
This project is funded through NASA’s Biodiversity Ecology and Conservation Program.